RDKit-derived features#
Via the RDKitAdaptor class you can wrap any featurizer that works on RDKit molecules into a featurizer that operates on BUS.
As an example, see how some of the featurizers below are implemented.
from rdkit.Chem.Descriptors3D import InertialShapeFactor as InertialShapeFactor_RDKIT
class InertialShapeFactor(RDKitAdaptor):
"""Featurizer for the RDKit InertialShapeFactor descriptor."""
def __init__(self):
"""Construct a new InertialShapeFactor featurizer."""
super().__init__(InertialShapeFactor_RDKIT, ["inertial_shape_factor"])
def citations(self) -> List[str]:
return self.super().citations() + [
"@incollection{Todeschini2008,"
"doi = {10.1002/9783527618279.ch37},"
"url = {https://doi.org/10.1002/9783527618279.ch37},"
"year = {2008},"
"month = may,"
"publisher = {Wiley-{VCH} Verlag {GmbH}},"
"pages = {1004--1033},"
"author = {Roberto Todeschini and Viviana Consonni},"
"title = {Descriptors from Molecular Geometry},"
"booktitle = {Handbook of Chemoinformatics}"
"}"
]
Instead of subclassing, you can also simply use the following syntax
from mofdscribe.bu.rdkitadaptor import RDKitAdaptor
from rdkit.Chem.Descriptors3D import InertialShapeFactor
my_featurizer = RDKitAdaptor(InertialShapeFactor, ["inertial_shape_factor"])
Ligand shape#
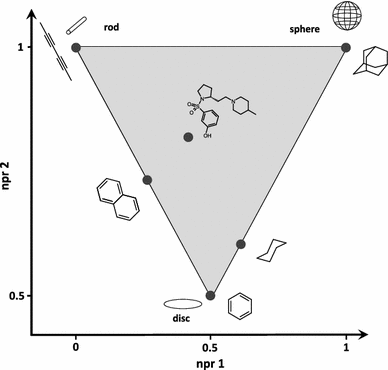
This descriptor is computed as NPR1+NPR2-1, and has been proposed by [Wirth]. |
This descriptor is computed as 2 - 2 * NPR2, and has been proposed by [Wirth]. |
This descriptor is computed as NPR2 - NPR1, and has been proposed by [Wirth]. |
Direct RDKit ports#
The following featurizers are the wrapped RDKit implementations (under the same name).